
As part of the Discovery Hub within The Royal Society’s Summer Sessions outreach event, all members of the Sanchez-Baracaldo Lab were showcasing the amazing world of microbes through a range of outreach activities. The Royal Society Summer Sessions event is the ideal opportunity for researchers from a wide range of disciplines to engage with the public in the work they do, and helps show the important part science plays in improving our quality of life, through solving some of the pressing issues facing the world today such as food availability, antimicrobial resistance and climate change.
Our lab focuses on cyanobacteria and diatoms, aiming to understand the biogeochemical processes in which they are involved, as well as the evolution of specific features they possess. Examples include showing how specific enzymes may have evolved, or how certain bacteria became tolerant to habitats of differing salinity. The answer to these questions involves working with the DNA of these organisms, with techniques such as genome extraction and sequencing, comparative genomics to observe differences in genomes and molecular clocks to estimate the geological time period over which bacterial traits arose.
Our first outreach activity was to show part of the process in DNA replication, members of the public could try their hand at being DNA Polymerase through the videogame “DNA PolymeRACE” where players must match DNA bases together, with speed and accuracy being key in achieving a high score.

Visitors were also shown how easy it can be to make models of DNA using only sweets! With strawberry laces for the sugar-phosphate backbone and four different coloured jelly-babies to represent the four DNA nucleobases Thymine, Adenine, Guanine and Cytosine.
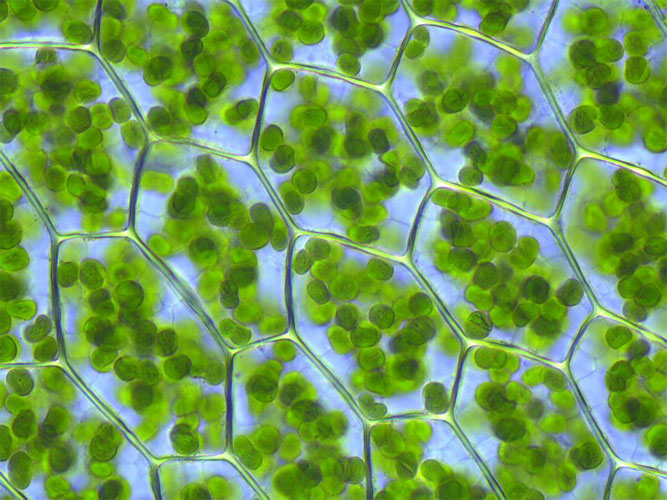
The lab also brought along some examples of our current Cyanobacteria and Diatom cultures, along with photographs to demonstrate the diverse range of morphologies. Examples of the lab cultures were also brought along fixed to microscope slides, so guests could get a detailed view of the cells the lab group work with.

Finally, to demonstrate the unique habitat of frozen sea ice, example blocks of sea ice and freshwater ice were brought along. Using yellow dye guests could see how sea ice provides a unique habitat for ice dwelling microbes such as bacteria and diatoms. This is due to how sea ice features numerous small spaces for these organisms to grow in comparison to freshwater ice, giving an insight into the earth’s polar regions.
Outreach events such as this are the perfect opportunity to showcase the science our lab undertakes as well as the big questions we wish to tackle. It’s also a great chance for us to see the other exciting work researchers within the Royal Society are carrying out.











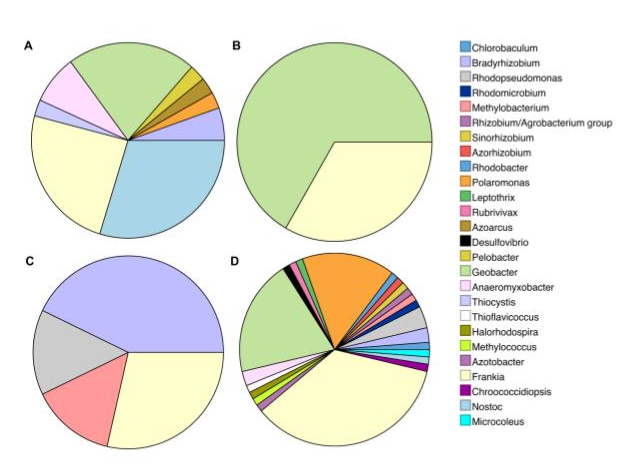
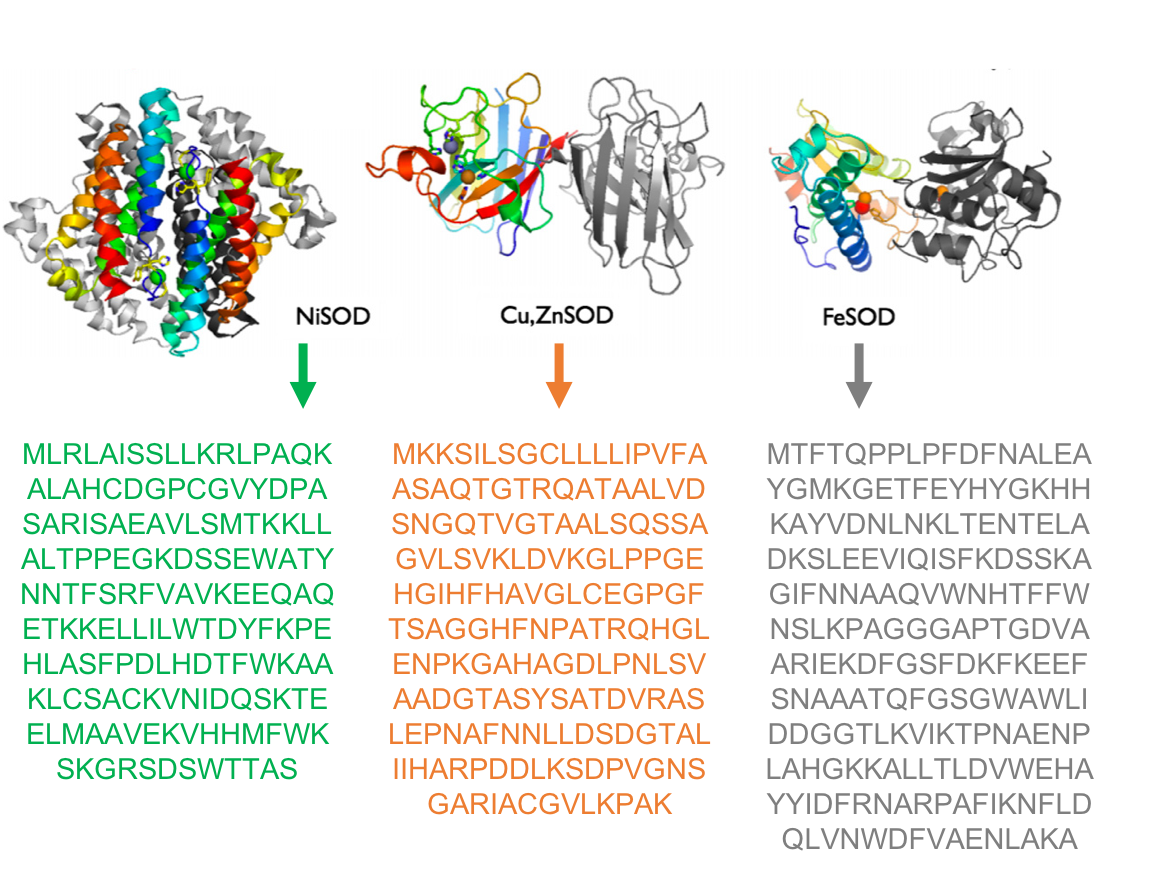
 Scientists have known that algae and land plants evolved after a more complex organism with a nucleus known as eukaryotes; an ancient eukaryote swallowed a cyanobacterium. While, it is accepted that cyanobacteria, are the ancestors of the chloroplast, it is unclear which of the cyanobacteria are closest related to the chloroplast, when this association first appeared in geological terms, and in which type of habitat this association first took place.
Scientists have known that algae and land plants evolved after a more complex organism with a nucleus known as eukaryotes; an ancient eukaryote swallowed a cyanobacterium. While, it is accepted that cyanobacteria, are the ancestors of the chloroplast, it is unclear which of the cyanobacteria are closest related to the chloroplast, when this association first appeared in geological terms, and in which type of habitat this association first took place.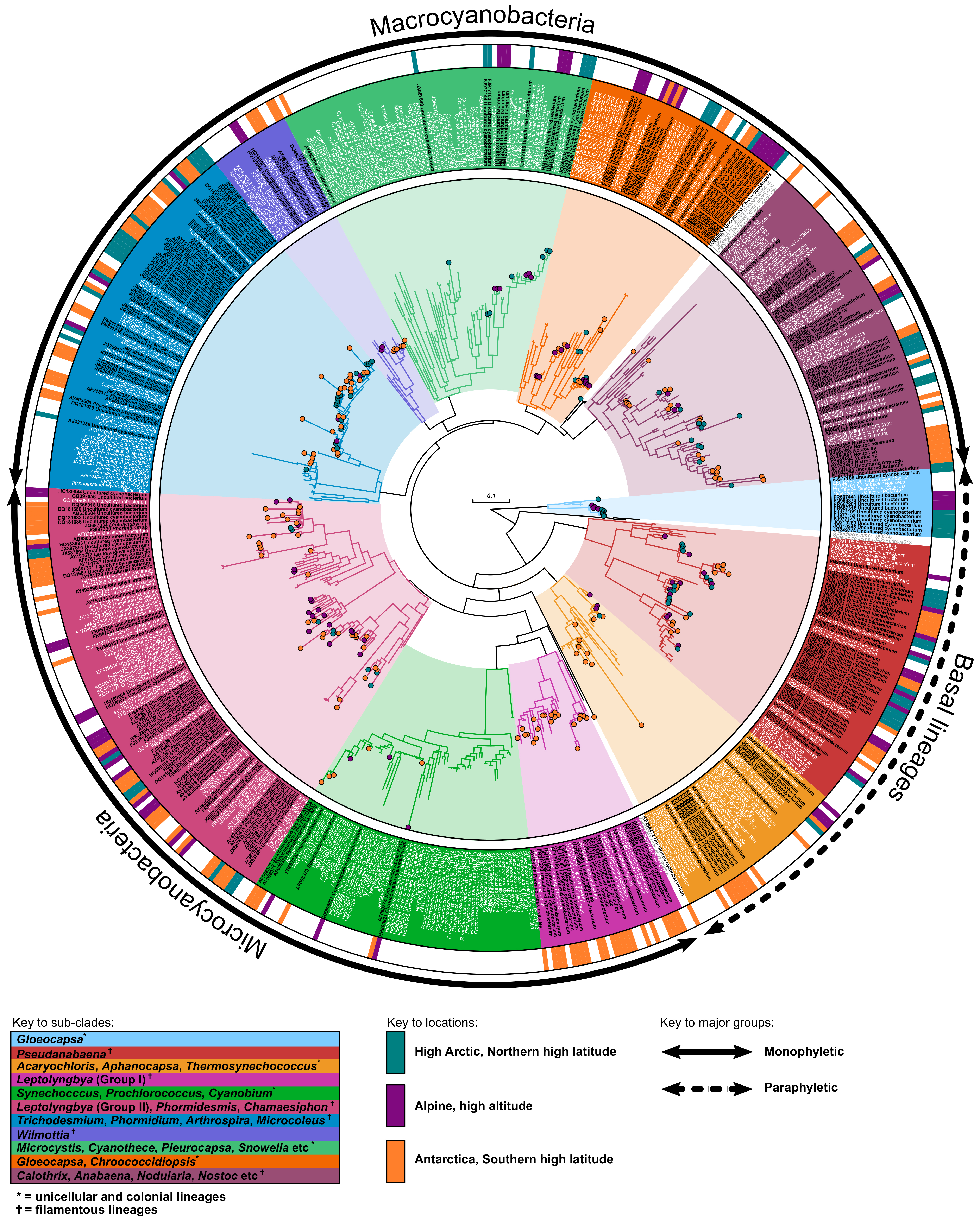 Nathan’s gets top price for figure published in our paper ‘
Nathan’s gets top price for figure published in our paper ‘